Managing Risks from AI-Enabled Biological Tools
Some experts are concerned that AI chatbots could pose biological risks. But AI-enabled “biological tools” may pose greater risks and will likely require a distinct policy response.
John Halstead
GovAI research blog posts represent the views of their authors, rather than the views of the organisation.
Introduction
Some experts have recently expressed concern that AI models could increase the risk of a deliberate bioweapon attack. Attention has primarily focused on the controversial hypothesis that “large language models” (LLMs) — particularly chatbots such as ChatGPT and Claude — could help scientific novices learn how to identify, create, and release a pandemic-causing virus.
The risks posed by AI-enabled biological tools have received less attention. However, some of these tools could pose major risks of their own. Rather than lowering skill barriers to misuse, some biological tools could advance the scientific frontier and make more deadly attacks possible.
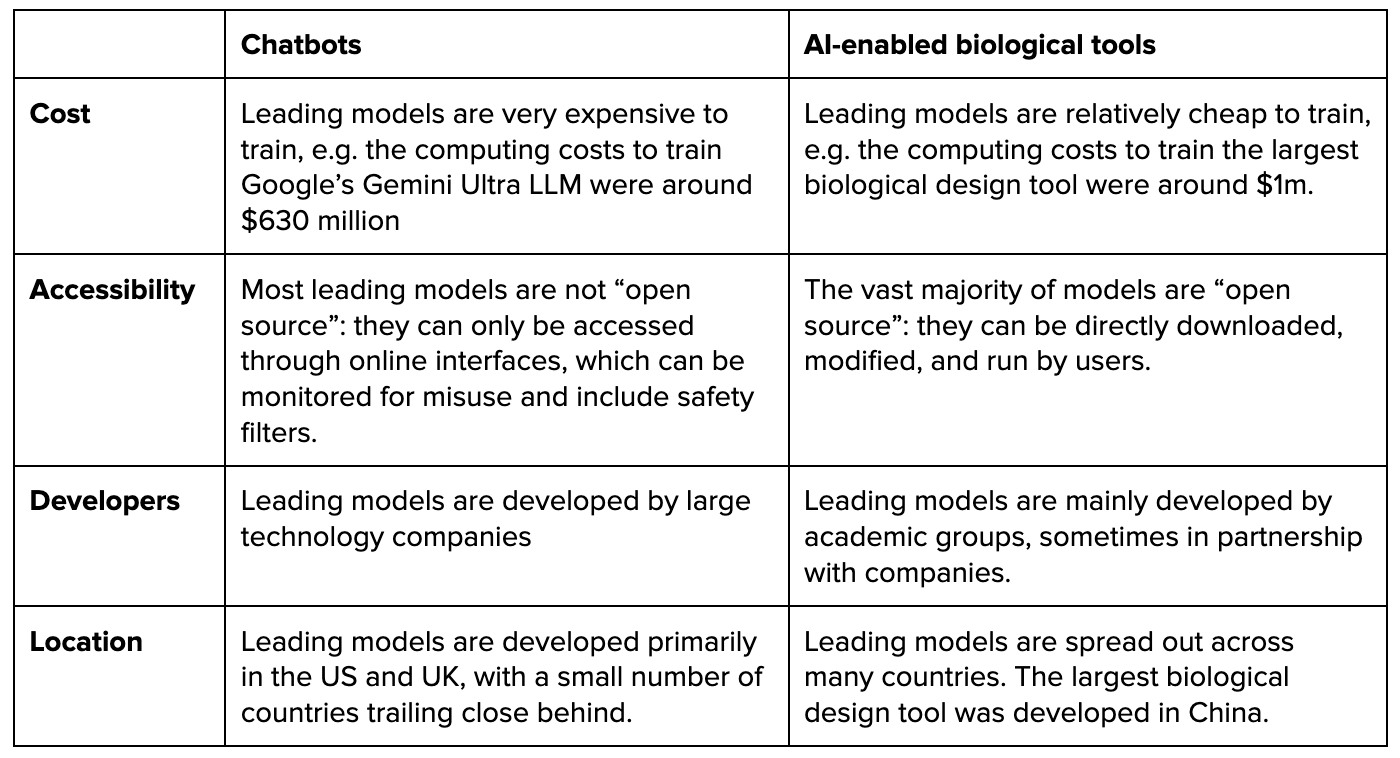
Unfortunately, policies designed primarily with chatbots in mind may have limited effects in addressing risks from biological tools. For example, policymakers focused on chatbots have begun to consider or enact mandatory reporting requirements for models trained using very large amounts of compute. The justification for limiting these requirements to high-compute chatbots is that compute usage strongly predicts the capabilities and thus risks of chatbots. However, for AI-enabled biological tools, it is an open question how far compute is now, and will be in the future, a good proxy for model capability and risk. Therefore, reporting requirements based on “compute thresholds” may not be effective at identifying high-risk biological tools.
Moreover, while cutting-edge LLMs are produced by only a handful of technology companies, biological tools are produced by a fairly large number of research groups that are dispersed across multiple jurisdictions. This makes self-regulation less likely to succeed. It also increases challenges around regulatory compliance and may create a need for different regulatory strategies.
Finally, policymakers focused on risks from chatbots have advocated for “red-teaming” as a method of identifying risks: this means trying to elicit unacceptable outputs from chatbots, such as advice about how to build biological weapons. However, red-teaming is less likely to be a practical and safe approach to assessing the risks of biological tools. For example, it would likely be both too dangerous and too costly to try to use a biological tool to design a novel virus and then test its virulence. In general, trying to produce new scientific knowledge about biological weapons is more problematic than trying to reproduce existing knowledge.
It is not yet clear what the most effective policy approach to managing risks from biological tools will be. However, a natural first step would be for governments to conduct recurring literature-based risk assessments of biological tools. This would involve surveying the literature for published evidence of dangerous capabilities in existing tools. The results of these assessments could inform policymakers in choosing which further tests to conduct, as well as when and whether new policy actions are warranted.
Moreover, there are some actions policymakers could take to mitigate both existing biological risks and potential new risks created by biological tools. For example, mandatory screening of DNA synthesis orders could, in general, make it harder for malicious actors to gain access to bioweapons by placing orders to mail-order virus suppliers.
Biological tools
AI-enabled biological tools are AI systems that have been developed using biological data such as genetic sequences and trained using machine learning techniques in order to perform tasks that support biological research and engineering. Perhaps the most notable biological tools are design tools, which can help to design novel biological agents — including novel viruses. However, the category also includes a broader range of tools such as platforms that automate wet lab experiments. Some biological tools can perform a wide range of biological tasks, while others perform specific tasks, such as predicting the structure of proteins (like AlphaFold 2).
Like chatbots, biological tools have made extraordinary progress in recent years. They are already contributing to progress in many areas of biomedicine, including vaccine development and cancer therapy. However, the models also introduce risks. Of particular concern are certain kinds of biological tools that could potentially allow malicious actors to design and develop pathogens that are more transmissible or more deadly than known pathogens.
The landscape of biological tools is different to the landscape of AI chatbots in several ways.
Risks from biological tools
To understand potential risk from biological tools, it is useful to first consider the “baseline risk” of a bioweapon attack, and then consider how biological tools could increase this risk.
Baseline risk
Independent of progress in AI, the baseline risk of a bioweapon attack over the next ten years is concerning. Today, thousands of people are already able to create viruses from scratch without using AI tools. Moreover, accessibility is increasing exponentially: it now costs only $1,000 to create the virus that caused the 1918 influenza pandemic from scratch1. (Note, however, that 1918 pandemic influenza is unlikely to cause a pandemic today, due to pre-existing population immunity.) The price of gene synthesis declined by a factor of 1,000 from the year 2000 to 2016.
Biological tools, therefore, are not necessary for a bioweapons attack. The concern is that some biological tools could exacerbate the risk.
How could biological tools affect biological risk?
Many experts have expressed concern that some kinds of biological tools could help malicious actors to identify new pandemic-capable viruses. For example, certain tools could help an actor to predict a virus’ toxicity or predict whether it could evade a vaccine.
Most AI-enabled biological tools present limited risks. However, it is difficult to know the degree to which some tools could increase biological risks. While there has been research into how chatbots affect the risk of a bioweapons attack, there are still no published risk assessments of the risk posed by current or future biological tools. It is also likely that a large portion of work on this topic will remain unpublished or highly redacted, due to concerns about spreading knowledge that could be misused. These same concerns are why I have not provided more specific descriptions of how biological tools could be misused.
Who can make use of biological tools?
The risks from biological tools will depend, in part, on how many actors can make use of them.
At present, scientific novices are unlikely to use them successfully. Barriers to use include both scientific knowledge and programming expertise.
However, these barriers could be overcome by general-purpose AI systems that can interface with biological tools. These models could interpret plain-language requests, choose and program the appropriate biological tool, translate technical results back into plain language, and provide a step-by-step laboratory protocol or even control automated laboratory equipment to produce the desired result. This is analogous to the way in which general-purpose AI systems, like ChatGPT, can already write and run simple computer programs in response to plain-language requests from users. (See this video from 310 copilot which outlines how such a system could work.)
These kinds of general-purpose AI systems are not yet mature enough to empower a novice2. They have shown some limited success, though, and in future they could ultimately allow novices to make use of biological tools too.
Even if the models cannot yet empower novices, they may still increase risks by increasing the capabilities of sophisticated actors.
Policymakers have taken only limited steps to understand and govern risks from biological tools
So far, policymakers worldwide have taken only limited steps to understand and address potential risks from biological tools. (See Appendix A.)
In the US, the main step policymakers have taken is to require the developers of certain biological tools to report information about these tools if they are trained using an amount of compute that exceeds an established threshold. However, the relevant “compute threshold” is set high enough that it may currently apply to only a single existing model.
Meanwhile, in the UK, policymakers have not yet established any regulatory requirements targeting biological tools.
Even though the EU has recently passed a wide-ranging AI Act, the Act does not cover AI-enabled biological tools.
Distinct governance challenges
There is currently a lively policy dialogue about how to assess and manage potential extreme risks from AI, including biological risks.
This dialogue has mostly focused on risks from chatbots and other language models. In this context, common policy proposals include:
- Supporting industry self-regulation by working closely with large technology companies to encourage them to voluntarily commit to responsible practices
- Encouraging or requiring the developers of models to red team them (i.e. test their safety by directly trying to elicit unacceptable outputs) and then put in place safeguards if sufficiently dangerous capabilities are found
- Creating standards or regulations that specifically apply to AI systems that exceed some compute threshold (since compute is highly predictive of the capabilities of language models)
However, because biological tools pose distinctive governance challenges, policy ideas that are developed with chatbots in mind often do not straightforwardly apply to biological tools.
Some distinctive challenges for the governance of biological tools are:
- Lower barriers to development: Leading biological tools are relatively cheap to develop, and developers are spread across numerous institutions and countries. Moreover, as the cost of compute declines and algorithms improve, the set of actors who have the resources necessary to develop biological tools will increase rapidly.
This could make effective self-regulation difficult to sustain: a large and heterogeneous set of developers — rather than a fairly small and homogenous set of technology companies — would all need to voluntarily follow the same norms. This could make it harder to ensure compliance with regulations. Further, it increases the risk that developers would move jurisdiction in order to avoid regulation.
- Greater risks and costs of red-teaming: Red-teaming biological tools (i.e. testing their safety by trying to elicit and identify unacceptable outputs) may be both more dangerous and more difficult than red-teaming chatbots. For the most part, red-teaming efforts that focus on chatbots attempt to assess whether these chatbots can help non-experts gain access to or apply dangerous information that experts already possess. On the other hand, red-teaming efforts that focus on biological tools may need to assess whether the tools can produce new dangerous information (e.g. a design for a new deadly virus) that was previously unknown. Clearly, in this latter case, the potential for harm is greater. Furthermore, actually judging the validity of this new information could require biological experiments (e.g. synthesising and studying a potentially deadly new virus) that would be both difficult and highly risky.
Therefore, policymakers will likely need to be more cautious about calling on the developers of biological tools to engage in red-teaming. Many developers of biological tools have themselves noted that evaluations “should be undertaken with precaution to avoid creating roadmaps for misuse”.
- Stronger open science norms: Unlike with cutting-edge chatbots, the vast majority of cutting edge biological tools are “open source”: they can be directly downloaded, modified, and run by users. This matters because safeguards against misuse can generally be easily and cheaply removed from any model that is open-sourced.
The commitment to open sourcing in the field in part stems from strong open science norms in the biomedical sciences in general and among biological tool developers in particular. Even when models are not open-source, these norms may also lead well-intentioned scientists to publish papers that call attention to information produced by biological tools that could be misused.
The upshot is that calls for biological tool developers to introduce safeguards against the misuse of their models could have comparatively little efficacy. If policymakers ultimately judge that certain safeguards should be put in place, they will likely need to rely especially strongly on regulatory requirements.
- Greater difficulty identifying high-risk models: In part because biological tools are more heterogeneous than language models, we should expect there to be a less reliable relationship between the level of compute used to develop a biological tool and the level of risk it poses. This means that “compute thresholds” may also be less reliable for distinguishing high-risk biological tools from low-risk ones.
If standard-setting bodies or regulators want to apply certain requirements only to “high-risk” biological tools, they may need to go substantially beyond simple compute thresholds to define the high-risk category.
In addition to these distinctive challenges, the fact that biological tools are also dual-use — meaning that, like language models, they can be used for good or ill — will tend to make governance especially difficult. It will be difficult to reduce risks from misuse without also, to some extent, interfering with benign uses.
Policy options
Despite all of these challenges, there are steps that governments can take today to understand and begin to mitigate potential risks from biological tools.
Two promising initial steps could be to (1) carry out recurring domain-wide risk assessments of biological tools, and (2) mandate screening of all orders to gene synthesis companies.
Conducting literature-based risk assessments of biological tools
In order to manage the risks of biological tools, governments should first review the published literature on models that have already been released to assess the risks the models pose. This would allow governments to understand whether and how to take further action.
As noted above, red-teaming models — or asking developers to red-team their own models — could involve unacceptable risks and unreasonable costs. Fortunately, as has also been noted by researchers at the Centre for Long-Term Resilience, conducting risk assessments that are based simply on reviews of published literature would be much safer and more practicable.
In slightly more detail, governments could proceed as follows. First, they could develop a list of potentially dangerous model capabilities and assessments of how much risk would be created by tools with these capabilities. Then, they could review the published literature from industry and academia to assess the extent to which already-released models appear to possess these capabilities.
These literature-based risk assessments would not substantially increase biosecurity risks, because they would not seek to discover new information that has not already been published. (However, because they could still draw attention to dangerous information, they should not be shared publicly.) Performing these risk assessments also would not involve running costly experiments.
Literature-based risk assessments would be an important first step in understanding and managing the risks of biological tools. They would provide governments with up-to-date information on the risks posed by current biological tools. This would in turn help governments to decide whether further actions are warranted. Possible responses to concerning findings could include: conducting more thorough assessments; supporting additional research on safeguards; issuing voluntary guidance to biological tool developers; or introducing new regulatory requirements. It is possible, for example, that there will eventually be a need for a regulatory regime that requires developers to seek government approval before publishing especially high-risk tools. Literature-based risk assessments could help governments know if and when more restrictive policy is needed.
Mandate gene synthesis screening
There are a range of policies that would reduce the specific risks from biological tools, as well as other biological risks that are not caused by AI. One such policy, supported both by many biosecurity experts and many biological tool developers, is gene synthesis screening.
Once many companies and scientists today have the DNA sequence of a virus, they can create synthetic versions of that virus from scratch. Some, though not all, of these gene synthesis companies screen orders from customers so that they will not synthesise dangerous pathogens, such as anthrax or smallpox. In the US, the White House recently developed best practice guidelines for DNA synthesis providers, which recommend that synthesis companies should implement comprehensive and verifiable screening of their orders. All federal research funding agencies will also require recipients of federal research funds to procure synthetic nucleic acids only from providers that implement these best practices. This is a valuable first step towards comprehensive DNA synthesis screening, but significant gaps remain. DNA screening should be mandatory for all virus research, including research funded by the private sector or foundations.
In the future, biological design tools may make it possible for malicious actors to bypass DNA screening by designing agents that do not resemble the function or sequence of any known pathogen. Screening requirements would then have to be improved in tandem with biological design tools in order to reduce this risk.
Conclusion
Rapid improvements in biological tools promise large benefits for public health, but also introduce novel risks. Despite that, there has so far been limited discussion of how to manage these emerging risks. Policy approaches that are well-suited to frontier AI chatbots are unlikely to be effective for mitigating risks from biological tools. A promising first step towards effective risk management of biological tools would be for governments to mandate screening of all gene synthesis orders and to carry out recurring risk assessments of released models that rely on the published literature.
Acknowledgements
Thank you to my colleagues at GovAI and to Jakob Grabaak, Friederike Grosse-Holz, David Manheim, Richard Moulange, Cassidy Nelson, Aidan O’Gara, Jaime Sevilla, and James Smith for their comments on earlier drafts. The views expressed and any mistakes are my own.
John Halstead can be contacted at john.halstead@governance.ai
Appendix A. Policy activity in the US, UK, and EU
As noted above, policymakers have taken only limited steps to understand and address potential risks from biological tools. Here, in somewhat more detail, I will briefly review the most relevant policy developments in the US, UK, and EU.
United States
In the US, there are no regulations that require the developers of any biological tools to assess the risks posed by their tools – or to refrain from releasing them if they appear to be excessively dangerous.
However, a provision in the recent Executive Order on AI does place reporting requirements on the developers of certain biological tools. Specifically, these requirements apply to anyone developing an AI model that is trained “primarily on biological sequence data” and uses a sufficiently large amount of compute. The developers of these models must detail their development plans and risk-management strategies, including any efforts to identify dangerous capabilities and safeguard their models against theft.
In practice, currently, these reporting requirements apply to very few biological tools. At the time of writing, only three publicly known models — xTrimoPGLM-100B, ESM3, and Galactica — appear to exceed the relevant compute threshold (1023 “floating point operations”). However, the number of tools that the requirements apply to is also likely to grow over time.
United Kingdom
The UK has not yet introduced any regulatory requirements (including reporting requirements) that target biological tools.
However, it has recently established an AI Safety Institute to support the scientific evaluation and management of a wide range of risks posed by AI. While the institute primarily focuses on general-purpose AI systems, it also currently supports work on risks from AI-enabled biological tools. It has not yet reported substantial information about this workstream, but may report more as the workstream matures.
European Union
The EU has recently passed an AI Act that will introduce a wide range of new regulatory requirements related to AI. The act places requirements and restrictions on (1) specific applications of AI systems, including “high-risk” AI systems; and (2) general-purpose AI systems. AI-enabled biological tools are not currently included in either category. This can only be changed by amending legislation, which involves the European Council, Parliament, and Commission.
General-purpose models
General-purpose AI models are defined according to “the generality and the capability to competently perform a wide range of distinct tasks” (Article 3(63)). This is not yet precisely defined, and it is unclear whether it could apply to AI-enabled biological tools. Some AI-enabled biological tools perform a wide range of biological tasks, but still perform a much narrower range of tasks than models like GPT-4 and Claude 3. Some potentially risky AI-enabled biological tools do not perform a wide range of biological tasks, and therefore cannot be defined as general-purpose.
The act says that a general-purpose AI model shall be presumed to have “high impact capabilities” and thus “systemic risk” if (but not only if) it is trained on more than 1025 FLOPs (Article 51(2), 51(1)(a))3. This does not apply to any existing AI-enabled biological tools (Maug et al 2024).
However, as noted in the main piece, in the future, it is likely that there will be combined LLM and biological tool models that are general-purpose in the sense meant by the AI Act, and will have advanced biological capabilities that may significantly increase biological risk. These models would be covered by the Act.
High-risk applications
Article 6 of the EU AI Act defines “high-risk” AI systems as those that “pose a significant risk of harm to the health, safety or fundamental rights of natural persons”. Annex III of the Act sets out specific criteria which classify AI systems as high-risk if they are intended to be used in high-risk applications, including use in social scoring systems, making employment-related decisions, assessing people’s financial creditworthiness, and other criteria.
It seems unlikely that AI-enabled Biological Tools could be classed as high-risk, for two reasons. First, the criteria set out in Annex III refer to how AI systems are intended to be used, but developers do not intend for Biological Tools to be used for biological weapons. Second, increased biological weapons capabilities are not included in Annex III. The European Commission can class systems as high-risk or not high-risk, provided they increase risk in the areas covered by Annex III (per Article 6). The Commission cannot use delegated acts to add new areas of risk that are not already included in Annex III (per Article 7). However, the Commission can propose the addition of new risk areas to Annex III of the Act to the European Parliament and Council once a year (per Article 112).
Scientific R&D exemption?
The Act states that AI models developed and used for the sole purpose of scientific R&D do not fall within the scope of the act (Article 2(8)), and this could plausibly apply to AI-enabled biological tools. However, since most AI-enabled biological tools are open source, there is no way to know whether they are being used solely for the purpose of scientific R&D, or are also being misused for bioweapons development by malicious actors. So, it is not clear that this provision does exempt AI-enabled biological tools from coverage by the Act.
Appendix B. Limits to the scaling of AI-enabled biological tools
AI-enabled biological tools have scaled rapidly over the past decade. To predict how the capabilities of biological tools will evolve, it is helpful to consider the different drivers of performance improvements. We can think of these improvements as being driven by (1) increases in computing power (or “compute”); (2) by increases in data quantity and quality; and (3) by algorithmic progress.
Here, I perform a preliminary analysis of how compute and data are likely to develop in the near-term. By my own assessment, the compute and data used in training is likely to increase rapidly in the near future and then increase more slowly thereafter. More research on this question would be valuable, as it would provide a clearer picture for policymakers of how risks are likely to develop in the future.
However, it is important to note that for AI-enabled biological tools, the sheer amount of compute and data used in training may not be a good predictor of especially risky capabilities. There may be models trained with small amounts of compute and data that provide significant uplift at specific narrow tasks that in turn materially increase misuse risk. Therefore, although the increase in compute and data used in training may soon slow, this does not necessarily mean that the increase in risks will slow proportionately.
Moreover, the analysis here focuses only on compute and data, but not algorithmic progress. More research on (a) the past effect of algorithmic improvements on the performance of biological tools, and on (b) likely future trends in algorithmic progress, would also be valuable.
Compute trends
The amount of compute used to train biological sequence models has increased by an average of 8-10 times per year over the last six years. This is equivalent to compute doubling around every four months. xTrimoPGLM-100B, a 100 billion parameter protein language model, exceeds the White House Executive Order’s reporting threshold of 1023 FLOP by a factor of six.
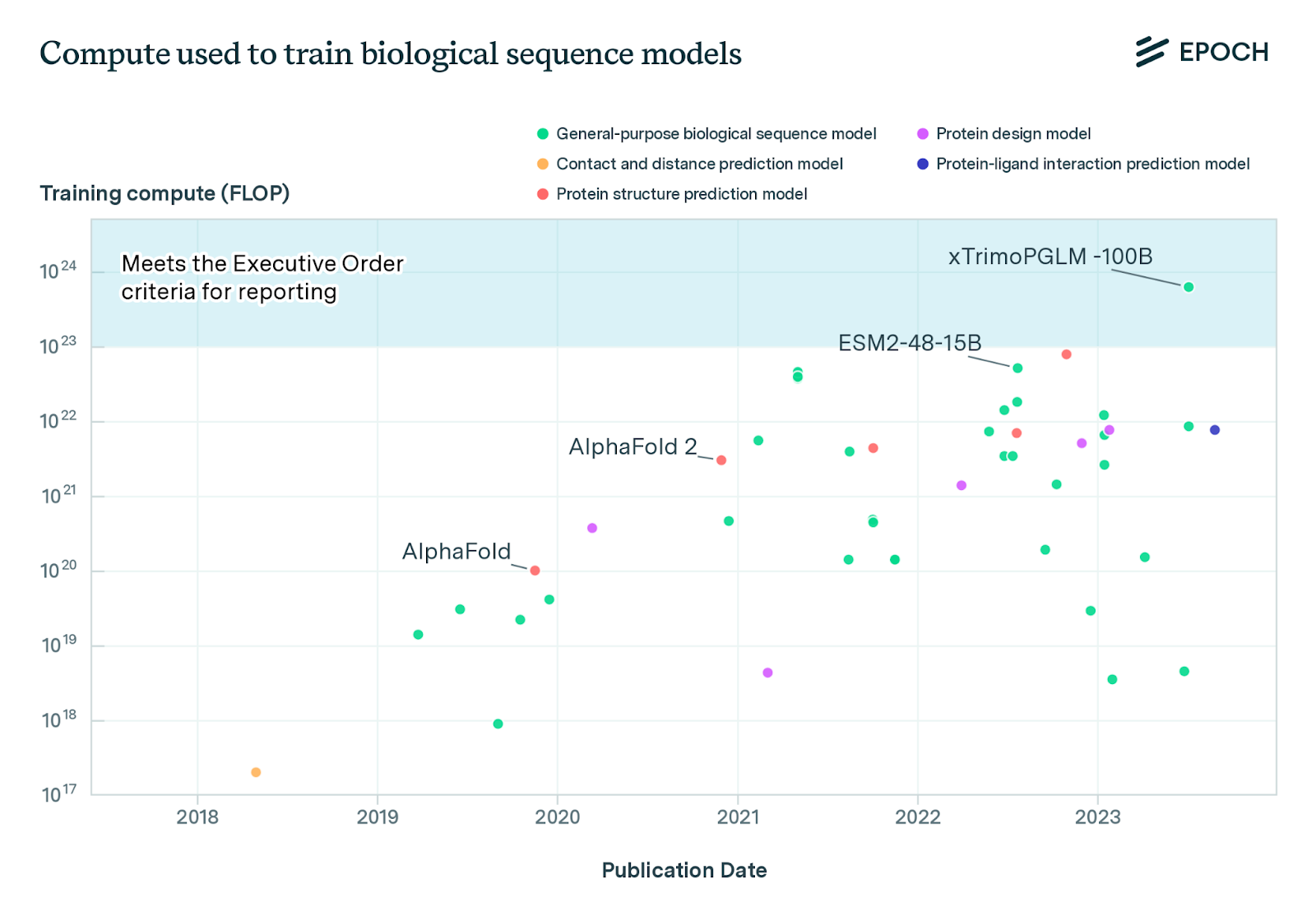
xTrimoPGLM-100B had amortised hardware training costs of around $1 million4. If biological design tools keep scaling at current rates, then the hardware cost of training will be on the order of $400 million in 2026 and $2.7 billion in 20275. Such costs would require biological design tools to have very large commercial benefits or receive substantial government funding. For comparison, it cost $4.75 billion over ten years to build the Large Hadron Collider at CERN. This suggests that the rate of increase in compute will decline substantially in the next few years. Future research should analyse likely future investment in biological tools from the public and private sector.
Data trends
There is much less information on trends in the amount of data used to train biological design tools than for compute. On the (questionable) assumption that the relationship between data scaling and compute scaling is similar to AI language models, then this would imply that the data used to train biological design tools is increasing by around a factor 13 per year6.
A report by Epoch found that the public databases used to train different types of biological design tools are increasing in size: between 2022 and 2023 key databases grew by 31% for DNA sequences, 20% for metagenomic protein sequences, and 6.5% for protein structures. If we assume that growth continues at this rate, and that all of this data is equally useful for model training, on current scaling trends, models trained on public databases cannot continue to scale at current rates beyond the next few years.7
However, this does not provide a realistic picture of the data constraints to scaling of biological tools for two reasons. First, the rate of the increase in data in public and private databases will likely increase in the future. Gene sequencing costs have declined by more than a factor of 100,000 over the last 20 years, and so are likely to decline by a few more orders of magnitude over the next few years. Various projects also propose a massive increase in sequencing. For example, the pilot Nucleic Acid Observatory proposal to sequence wastewater at all US ports of entry would increase the growth rate in metagenomic sequences by a factor of 100.8 So, there could be changes in the growth rate of available data in the future.
Second, some proprietary databases are much larger than public databases. For example, Basecamp Research, a private company that develops biological databases and develops biological design tools, aims to create the world’s largest and most diverse protein database. Basecamp says that after only a few years of sampling, their proprietary databases have 4x more sequence diversity and 20x more genomic content than comparable public databases. Indeed, there will be stronger commercial incentives to improve biological databases given the capabilities of Biological Design Tools. Future research should analyse the implications of these large proprietary databases for future scaling of biological design tools.
There are two caveats to this. First, there are diminishing returns to the increases in data in the relevant databases because many of the genomes are very genetically similar to one another. For example, there are around 1,000 human genomes and 260,000 e-coli genomes in the NCBI database, but these genomes are very similar to one another. So, the training data is less varied and therefore less useful for biological tools than, e.g. for LLMs.
Finally, state-of-the-art protein language models that predict protein structure and function are pre-trained in a self-supervised way on tens of millions to billions of proteins.9 However, in order to predict rare or novel protein functions, which are especially concerning from a biological risk point of view, current models need to be fine tuned using labelled protein data. Historically, labelling of protein function had been imperfect, highly labour-intensive, and required synthesising decades of biological research. If labelling continues to be labour-intensive, the quantity of labelled data is unlikely to increase by an order of magnitude in the near future.10
However, this could also change in the future because AI tools may themselves be able to label proteins more accurately and efficiently. For example, Basecamp’s HiFin-NN AI model outperforms other annotation models and is able to rapidly annotate a large representative portion of the MGnify microbial protein database that was previously not annotated.
Footnotes
1 - Gene synthesis companies offer a price of around $0.07 per base pair, and influenza is made up of around 13,000 base pairs.
2 - A recent report by researchers at the Centre for Security and Emerging Technology notes that “In published studies, chatbots paired with [biological design tools] have either only provided instructions or accomplished individual steps in larger experimental workflows and still required biological expertise to finish, required extensive programming expertise, or focused on chemical synthesis tasks, which are relatively straightforward compared to biological experimentation and [biological design tool] use”.
3 - So, the 1025 FLOP compute training threshold is a sufficient but not necessary condition for a system to be classed as general-purpose.
4 - Calculations are available in the ‘Cost trends’ tab of this sheet.
5 - Calculations are available in the ‘Cost trends’ tab of this sheet.
6 - See the ‘Cost trends’ tab of this sheet.
7 - Calculations are available in the ‘Protein and DNA data’ tab of this sheet.
8 - The proposal sketched out here would generate in one year ~30x the total amount of publicly deposited metagenomic sequence data to date (9E16 bases vs 3E15 bases; annual new data is currently c. 1E15 bp, i.e. the proposed system would 100x annual growth). The full scale proposal (5000 sites, worldwide) would generate ~1000x the existing total (2.7E18 bases) in one year.
9 - For a review of protein language models, see (Notin et al 2024).
10 - This is discussed in Crook et al. (2024), which is currently unpublished.

